Figure 1 Computer Simulation
Protein folding requires massive calculation.
By combining the advancing areas of protein research and information technology, bioinformatics is a path to the ultimate goal of realizing new biological knowledge. This will allow a deeper understanding of disease and medication. At the current stage, the missions include nucleic acid analysis arrangement, protein sequences, protein structures, protein families, and biochemical reactions, etc.
The topics that require information and semiconductor research assistances include: sequence alignment, sequence analysis, comparative genomics, computational genetics, gene identification, protein structure analysis, gene micro-array chip analysis, molecular evolution, drug design, and other fields.
High-performance computing design equipment would fulfill massive calculation requirements
Sequencing is the basis of gene research. Presently, we rely mostly on imports for much of the equipments including software and hardware for gene sequencing, so the question lies in how genes sequencing-related local industries will develop. The high-performance equipments have two primary applications:
1. Gene searching and comparison: by inputting known DNA structures into a computer, a sample sequence can be identified as well as the relation between that sequence and different DNA.
2. Protein folding simulation and computer-aided drug design: protein recombination and expansion is the basis for disease-prevention and drug-design. As such, computer-aided design and algorithm design are extremely important.
Under the three primary directions of geometry, energy, and activity, we can discuss further on how to use minimal energy to achieve an improvement in molecular docking during the drug design process, an improvement in drug-design timeline, and a decrease in research and development costs.
An FPGA for gene calculation applied design
Nvidia’s CUDA platform utilizes the C language to control the GPU development environment, which is well-suited for parallel computing. Under CUDA architecture, DNA sequencing, short assembling sequences can play to the advantages of parallel computation, speeding up computation and generation of three-dimensional genomes.
Invitrogen and Active Motif are biotechnology companies that have cooperated to utilize FPGA-based analysis to create a new generation of gene sequencing data. TimeLogic is a biological computing system that utilizes one or more groups of PCI Express x 1 FPGAs on boards, which integrates with the FPGA accelerator and genomics algorithm to speed up computation speeds to more than 100 times of traditional CPUs when comparing between newly discovered data and established gene knowledge.
Under higher requirements for protein folding simulation calculation, researchers first accelerate bonding positions of geometric searching by using hereditary calculations; then energy is the focus, where the stability theory of Lyapunov functions are used to decrease the number of bonding positions in order to further enhance the effectiveness of molecular docking. At the same time, the lowest energy state is achieved through speeding up the molecular system by adjusting the weighted control points and knot vectors. Lastly, a variety of models of drug receptors are simulated in a computer, while utilizing the least energy to determine the smallest area of stable docking, close to minimal global energy state and the assessment of a variety of molecular activity.
In order to utilize the FPGA chip to achieve molecular computation, as well as correspond with the Lyapunov function to decrease molecular docking geometry and stability, we use the system architecture shown in Figure 2 to meet requirements of higher-level researchers. As Figure 2 shows, the primary function of a single-IC system is to provide a high-speed input of data through the use of 4 GIGE ethernet sockets. This provides the back-end FPGA with processing sequence alignment. The core of the hardware architecture is the double DDR2 SODIMM module’s Multi-port Controller. The key for an overall fast system computation lies in the design of the memory management unit and high-speed bus. In order to realize the all-important Linked List data structure in an FPGA during sequencing comparison, a Zero Bus Turnaround (ZBT) SRAM is used to store the Pointer. The two read-write subsystems should operate at the same time without interference with one-another, effectively forming a dynamic parallel processor.
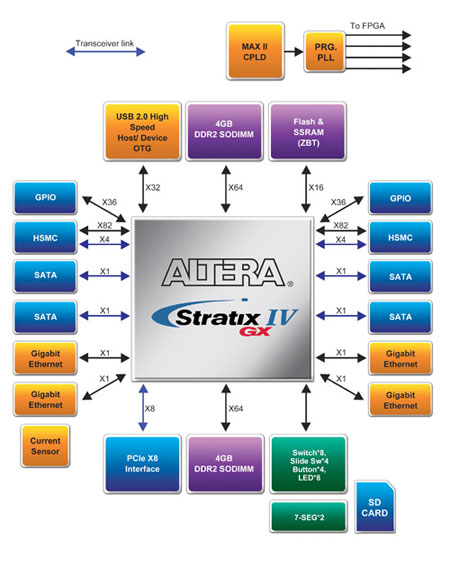
Figure 2 FPGA gene and protein calculation architecture
Regarding the sending and receiving data through DMA on the PC, the system uses a PCI Express x 8 to provide a 16 Gb/s bandwidth to meet requirements. The system also provides two HSMC connectors for high-speed interfacing, which provide up to 8 pairs of 10 Gb/s key bandwidth for high-speed signal lines.
Using HSPICE simulation to overcome 10Gb/s system design bottleneck
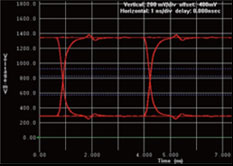
Figure 3 DDR2/DDR3 simulation results
The signal integrity of a high-speed FPGA development platform is the key to a successful system. Thus, in our designs, we utilize Hspice, which includes a variety of simulation tools. Through the analysis of specific issues to optimize the parts selection and design trade-offs, such as stacking structure, dielectric material, cable topology, cable length, width, and impedance matching components, as well as the design adjustments due to simulation results, most of the signal-integrity issues are solved within the time period of design.
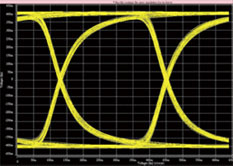
Figure 4 HSPICE eye-diagram result
Figure 5 shows the FPGA development platform made and based on the requirements of multi-party researchers. Currently, protein folding and drug-design experts in Taiwan utilize this platform to run accelerated algorithmic tests.
For more details, please visit de4.terasic.com
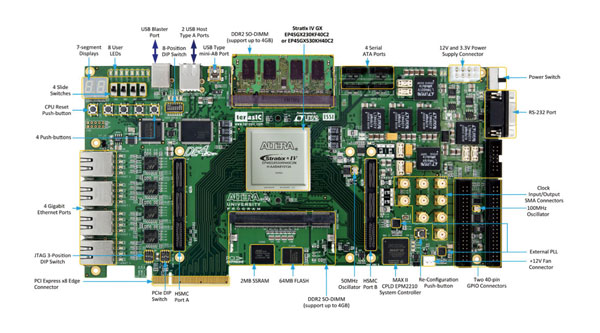
Figure 5 FPGA hardware platform
